#svm #machinelearning #ai
Note: The article is also on authors medium.com account in duplicate.
Here in this article, you will find SVM-based classification running codes for all kinds of SVM classifications. SVM (Support Vector Machines) are classification engines popularly known for 2-class classification since it separates data into 2-class by separating hyperplanes (See below the hyperplane separating colon cancer data, for an example). The support vectors provide the boundaries of the hyperplanes of the two classes. However, the two-class classification can be enhanced to classify multi-class classification problems as well using one versus all and one versus one approaches. SVMs can solve problems that are non-linearly separately by using Kernels and going in higher dimensions to find the separating planes between the two classes.
In this article let’s get directly into the problem of classification using Python on a well-known colon cancer dataset. The dataset is a binary class classification dataset, have cancer or don’t have cancer, are the two target classes.
There are various versions of SVM classifiers based on the kernel used. We assume you know what the kernel in SVM is. Here the classification problem is reduced to an optimization problem, which is solved using optimization techniques. The aim is to separate the two classes as far as possible, the hyperplane that does the same is chosen as the optimal separating hyperplane. With a Kernel the problem can be shifted to higher dimensions where the problem is linearly separable and a hyperplane can be found that separates data in higher dimensions. The kernel can be of various kinds, let’s start with the basic one.
With each kind of Kernel the output may change, here it is explained how to use each of the key kind of SVM.
Let us go through common steps for all kinds of SVM:
- Import modules, we are using the sklearn package from scikit-learn, lets start with “pip install scikit-learn”
- Read input file
- Define X and Y, as data and target class
- Split data in test and train using 0.3% as testing data
- Call SVM, one of the many SVM version you want to use
- Predict the output
- Compute Accuracy
I. Linear SVM
The following is the code when SVM used classification using a linear kernel.

The classification accuracy reached was 100%
Now let’s code for the RBF kernel
II. RBF kernel
The code for RBF Kernel is as follows, see the change in code at SVC parameters where kernel type is explicitly specified.

Here gamma is a parameter and can be changed for rbf kernels. The accuracy reduced to 68.42% for the RBF kernel, this shows the choice of the parameter was not optimal. RBF kernels have many advantages in solving non-linearly separable classification problems.
III. Polynomial Kernel
The code for Polynomial Kernel is as follows, see the change in code at SVC parameters where kernel type is explicitly specified.
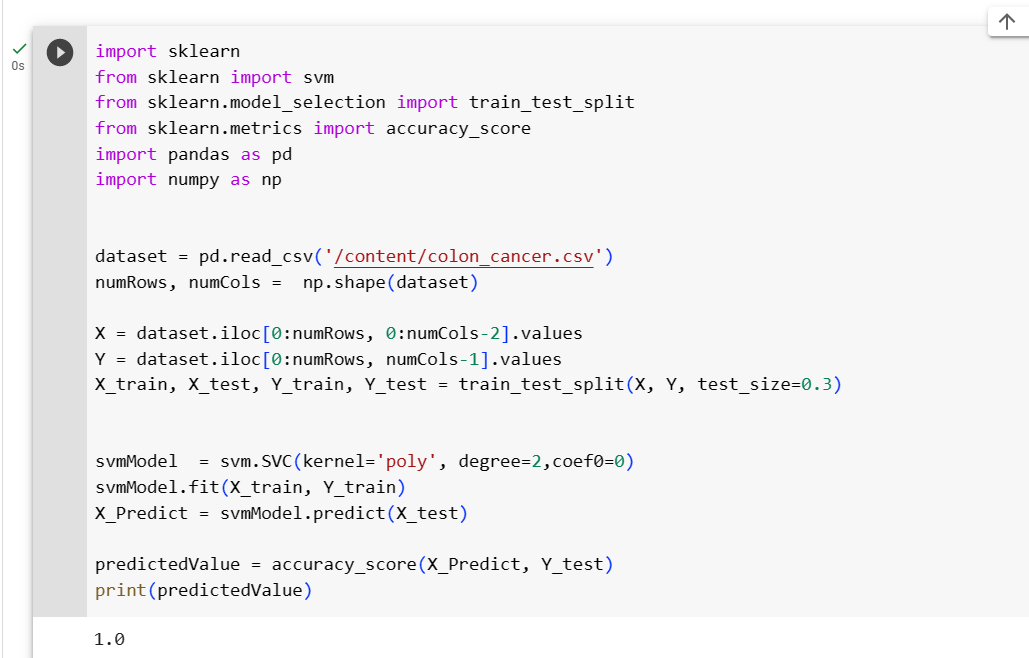
This is code for the second-degree polynomial kernel, specified in degree=2, and other polynomials which are optional can be tried as well. If it is not mentioned default values are taken for parameters. We reached an accuracy of 100%. Better!
IV. Sigmoid Kernel
The code for Sigmoid Kernel is as follows, see the change in code at SVC parameters where kernel type is explicitly specified.

The option of sigmoid kernel is filled in call to SVC and other optional parameter that can be missed is coef0, here its value is taken as 0. coef0 is optional, as its default value is taken if this is not specified. Accuracy reached again is 100%.
User-defined kernels can also be used in the same way.
This linearly separable data of colon cancer with support vectors, hyperplane, and boundaries can be viewed as follows in a plane:

These are all classification calls to SVM, many other SVM calls specific to multi-class classification problems can be performed too with slight changes.
The code in text format is here:
import sklearn
from sklearn import svm
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
import pandas as pd
import numpy as np
dataset = pd.read_csv('/content/colon_cancer.csv')
numRows, numCols = np.shape(dataset)
X = dataset.iloc[0:numRows, 0:numCols-2].values
Y = dataset.iloc[0:numRows, numCols-1].values
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.3)
svmModel = svm.SVC(kernel='sigmoid', coef0=0)
svmModel.fit(X_train, Y_train)
X_Predict = svmModel.predict(X_test)
predictedValue = accuracy_score(X_Predict, Y_test)
print(predictedValue)