#AI #artificialintelligence #ml #machinelearning
Note: This article is present on authors medium.com account as well.
In the past few articles, you might have got an idea of what feature selection is. In short, it is used to improve accuracy, decrease noise and at the same time decrease computational complexity by reducing irrelevant features from big datasets. Further, it informs about the importance of a feature as well.
The selection of features can be considered as a combinatorial optimization problem, where we are given n features, and for each feature, a decision is to be made to select or mark the feature as important or consider it as redundant, or a noise.
In this article, you shall learn how genetic algorithms can be used to perform feature selection. Here are the key points and this is followed by working code in python.
- The aim is to generate a population of candidates which can be evaluated for key features in the dataset.
- In each iteration of Genetic Algorithm (GA) new features are generated as part of GA, selection, and reproduction and mutation concepts. However, we are using python package, namely, geneticalgorithm. This library hides or encapsulates the inner working of geneticalgorithms and also hides the process of generation of the next candidate.
- This python library minimizes the objective function.
- We have taken objective function as accuracy generated by the system when a subset of features is chosen by GA is to participate in the classification problem.
- The final output of the code, is a subset of features that can be used in future data prediction of target class.
- The code is in python with explanations.
Python Implementation
Install the library
pip install geneticalgorithm
Import essential libraries
import pandas as pd
import numpy as np
import sklearn
from sklearn import svm
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from geneticalgorithm import geneticalgorithm as ga
Read the dataset
dataset = pd.read_csv('/content/colon_cancer.csv')
numRows, numCols = np.shape(dataset)
Define the objective function to optimize by Genetic Algorithm
def f(combinational_array):
print('the new ga array is: ')
print(combinational_array)
arrayToTest = getIndx(combinational_array)
print(arrayToTest)
X=dataset.iloc[0:numRows, arrayToTest].values
Y = dataset.iloc[0:numRows, numCols-1].values
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.3)
svmModel = svm.SVC()
svmModel.fit(X_train, Y_train)
X_Predict = svmModel.predict(X_test)
predictedValue = accuracy_score(X_Predict, Y_test)
print ("predictedValue: ")
print(predictedValue)
return -1 * predictedValue
Auxiliary function definition
def getIndx(arrayIndx):
lenArr = len(arrayIndx)
arrayOutput=np.zeros(lenArr)
j=0
for i in range(lenArr):
if arrayIndx[i] >= 0.5:
arrayOutput[j] = i
j=j+1
if j==0:
return ([0])
arrayReturn=np.zeros(j)
for i in range(j):
arrayReturn[i] = arrayOutput[i]
return arrayReturn
Define parameters and call the Genetic Algorithm to perform feature selection. The dimension of colon cancer data undertaken here is more than 1500, so let’s test on a smaller length of say 10 features. When computing over full data, increase the number of features to one-fourth of the number of columns at least to start with. 10 features are taken just as an example to show how it works.
length = 10
# length = int(numCols/4)
print("length is : ", length)
varbound=np.array([[0,1]]*length)
model=ga(function=f,dimension=length,variable_type='int', variable_boundaries=varbound)
model.run()
The sample output is as follows:
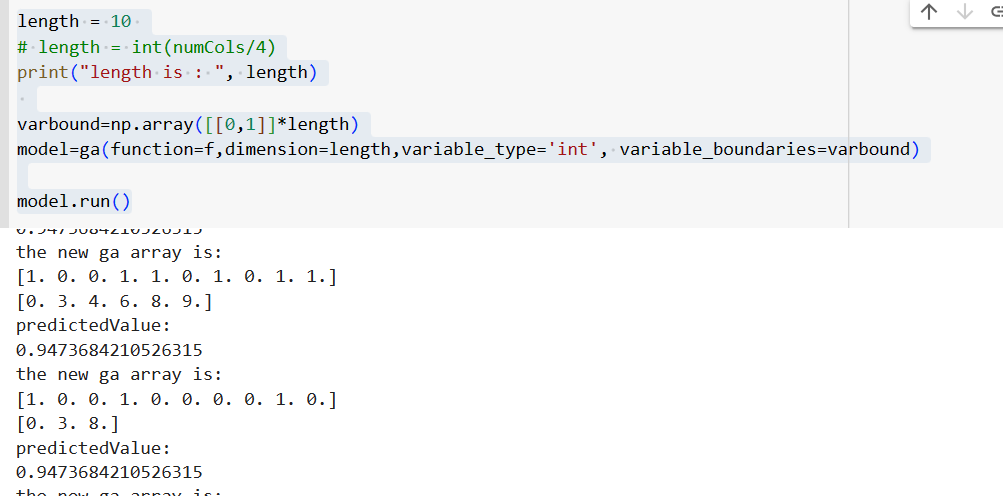
The sample output with more features is as follows:
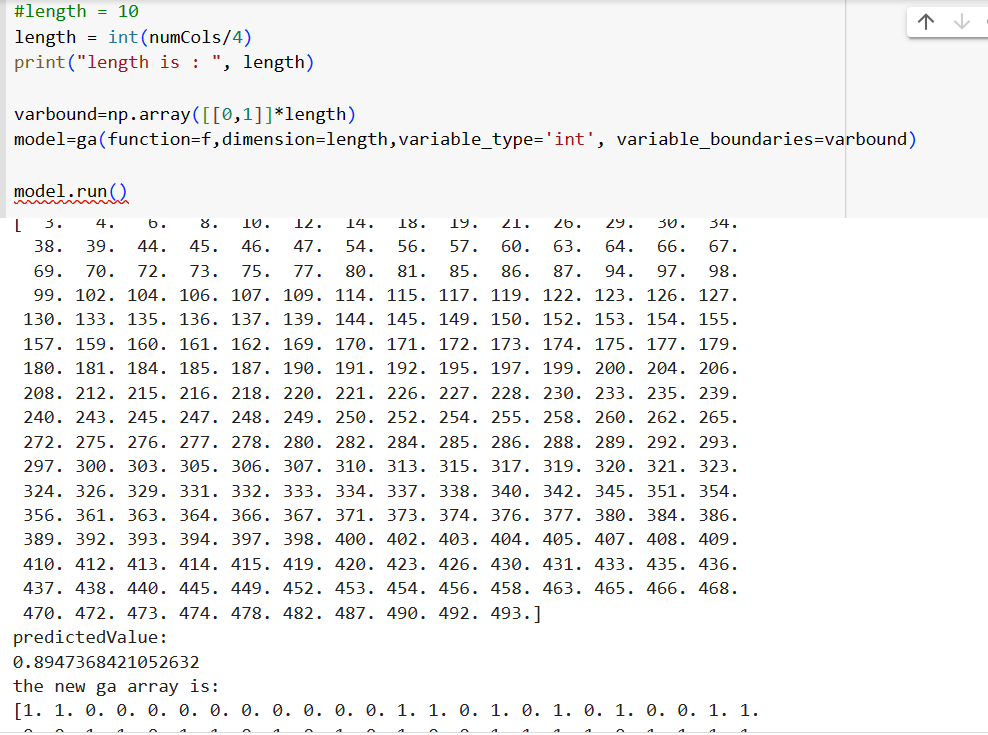
The aim is to choose both the number of features and the features themselves, both using inbuild GA based implementations.
Thank You for reading.
Interesting library I never found a good one in python for developing genetic algorithms I should look into it. Is there a way to also account for diversity of the population as well?
Most of the time I end up implementing my own. Here you can find a recent article that I wrote, your opinion is very welcome:
https://francescolelli.info/machine-learning/on-genetic-algorithms-as-an-optimization-technique-for-neural-networks/
LikeLike
Respected Dr. Francesco,
Yes this library was decent enough.
I could find only the basic functionalities of GA here. Still it is better than PSO library available in python. I wrote a blog on PSO with python as well.
Regarding your question about diversity of population I could not find transparency as of now. I shall try to look into code sooner to see if that can be integrated.
That’s so good that you implement your own code, as the world of opportunities open up with self implementation of code. This is because we know how to improve and how to hybridize the algorithm.
I have read your article very well written and knowledgeable articles it is.
Best Regards
Nidhika
LikeLiked by 1 person
Good evening Sir, here is the code for this library. https://github.com/rmsolgi/geneticalgorithm/tree/master/geneticalgorithm.
Yes there are options to customize the population diversity with mutations rate and selection. This can be done by providing the same in arguments to the called GA object. Hence, there is no need to go in code. However, when I get more time, I would like to get in code as well.
Here are parameters:
algorithm_param = {‘max_num_iteration’: None,\
‘population_size’:100,\
‘mutation_probability’:0.1,\
‘elit_ratio’: 0.01,\
‘crossover_probability’: 0.5,\
‘parents_portion’: 0.3,\
‘crossover_type’:’uniform’,\
‘max_iteration_without_improv’:None}
Best Regards
Nidhika
LikeLike